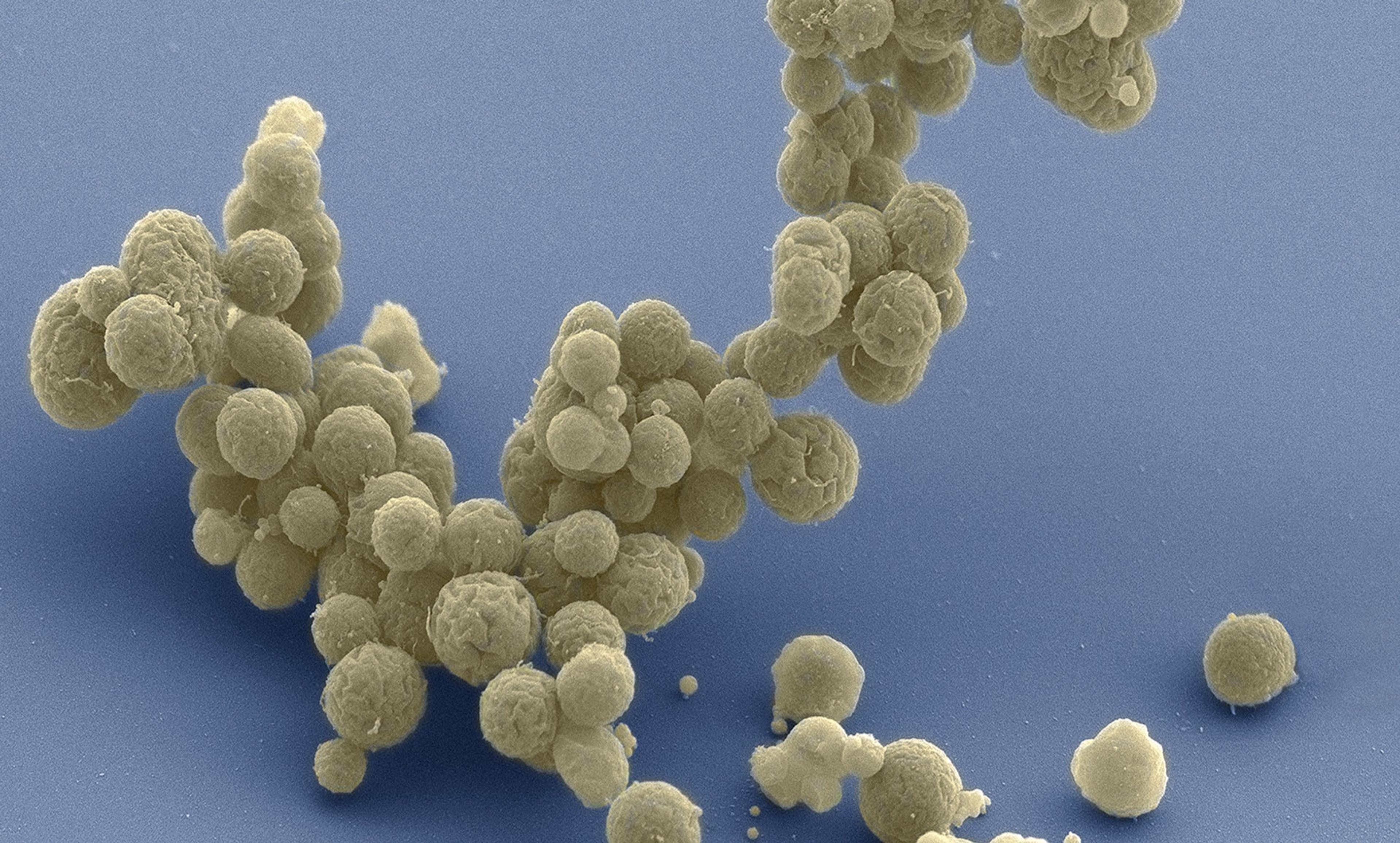
Courtesy Tom Deerinck and Mark Ellisman, National Center for Imaging and Microscopy Research, University of California at San Diego
The American physicist Richard Feynman once said: ‘What I cannot create, I do not understand.’ With that inspiration, my colleagues and I set out to attain a deeper understanding of life by assembling it ourselves. Over the past 15 years, our teams at the J Craig Venter Institute (JCVI) and Synthetic Genomics, Inc (SGI), both in La Jolla, California, have been developing tools to design whole genomes, synthesise and assemble them in the lab, and install them into a living cell. Our goal was not just to elucidate the genetic components required for life, but also to establish the capacity to create organisms custom-tailored to specific industrial applications. This spring we achieved a breakthrough in our work.
It hasn’t been easy. The technologies for chemically synthesising and activating whole bacterial chromosomes converged a full six years ago, when we announced the creation of the first synthetic cell. To build that cell, we first assembled 60-base single-stranded DNA (derived from the genome of the Mycoplasma mycoides yeast essentially as it occurs in nature) into double-stranded DNA fragments. We stitched these fragments together in the lab using a combination of biomolecules we discovered that can reassemble pieces of DNA. We then combined our new genetic sequences inside a yeast cell. The synthetic genome was 1,078,809 base pairs (genetic letters) long, the largest chemically defined structure synthesised in a laboratory.
In the final step, we carefully moved the complete synthetic genome out of the host yeast and transplanted it into a recipient cell. Similar to installing new software into a computer, the synthetic genome reprogrammed the recipient cell and converted it to the organism encoded by our rewritten genetic code. Since all of the properties of the cell were expressed from the synthetic genome, and the original traits of the recipient cell were diluted out as the cell grew and divided, we considered the new cell a synthetic cell, which we called Mycoplasma mycoides JCVI-syn1.0.
Over the past two decades, researchers have sequenced and stored thousands of complete bacterial genomes, but our syn1.0 synthetic cell was the first proof that we could reverse this process: we could pull a DNA sequence out of the computer, edit it, convert that revised sequence into a chemically synthesised structure, and create a free-living cell based on that new DNA sequence. Our syn1.0 work was only a first step, because we essentially started with the exact genome sequence from naturally occurring M mycoides, but it gave us an extraordinary set of tools for DNA construction and activation. Our work also established a design-build-test cycle for designing a whole bacterial genome.
Synthetic biologists aim to produce cells with new and improved biological functions that do not already exist in nature. Doing so requires a deep knowledge of what natural biology already does, but there is not a single self-replicating cell in which we understand the function of every one of its genes. We therefore have been working to create a synthetic minimal cell – one that has only the machinery necessary for life. And now we have succeeded.
In March we reported on the synthesis of a cell we call JCVI-syn3.0. It has the smallest genome of any cell that can autonomously replicate, with just 473 genes. JCVI-syn3.0 retains almost all of the known genes involved in the synthesis and processing of macromolecules and, surprisingly, 149 genes with unknown biological function. (Note that Feynman did not say: ‘What I can create, I do understand.’) Those genes highlight that our current knowledge of the genetic requirements for life is still limited. Our designs will remain restricted to naturally occurring DNA sequences until we can define the purpose of every gene and genetic element – including those mysterious 149.
The relative simplicity of syn3.0 and its well-characterised genome provides a starting point for studying how cells work at the most fundamental level. In the next phase of this project we will begin defining the function of every gene in syn3.0 so that we can model its behaviour on a computer. This cell will be an extremely useful chassis for learning about the first principles of cellular life and for discovering how to predictably impart new biological functions. We also imagine using the minimal cell to help us produce more complex microbial species, ones that could be valuable for industrial applications. A minimal cell will have several advantages in these applications. It would be devoting maximal energy to the product programmed into the cell. Because every gene is essential, a minimal cell would likely exhibit fewer cellular mutations. And because it is a simple system, it would be relatively straightforward to engineer.
Our synthetic cell work has been met with some worries about the potential and the safety of this level of genetic manipulation. As with any new area of science and technology, synthetic genomics has the potential for great societal benefit but also the potential for harm. Craig Venter and his teams at both JCVI and SGI have been publicly addressing the ethical and societal implications of synthetic life since we first proposed the creation of a minimal cell in 1999. JCVI’s Policy Group has helped inform federal agencies and the general public about ways to responsibly develop and regulate this new technology; so have others, such as those on President Obama’s Bioethics Commission. For example, our synthetic bacterial cells are designed so they cannot live outside of the lab or other production environments. They are dependent on certain specific nutrients without which they cannot survive.
Now that we have created syn3.0, we are working to predictably design and build organisms programmed to perform specific functions. We believe this technology will be essential for creating low-cost, environmentally sustainable industrial chemicals, medicines, biofuels and crops. We’ve already used our synthetic DNA technologies to stockpile an H7N9 vaccine in response to the 2013 H7N9 influenza outbreak in China. Over the next decade, we should start seeing a whole range of vaccines that are much more effective and produced much more rapidly. We also anticipate synthetic organisms being used to create pharmaceuticals tailored to an individual patient’s condition rather than produced en masse.
The possibilities of our technology are boundless. Clothes constructed from renewable, bio-based sources; cars running on biofuel from engineered microbes; plastics made from biodegradable polymers; customised therapies ‘printed’ at a patient’s bedside – these are just a few plausible benefits that could soon emerge from our effort to understand life by creating it.