Here is a remarkable fact about identical twins: they have the same DNA, and therefore the same ‘genetic fingerprint’, yet their actual fingerprints (such as they might leave behind on a murder weapon) are different, and can be told apart in standard police observations. Fingerprints are, of course, produced by the pattern of tiny ridges in skin. So, it would appear that certain fine-scale details of our anatomy cannot be determined by a precise ‘genetic blueprint’.
It isn’t only fine details that seem open to negotiation in this way: anyone who has seen Bonsai cultivation knows how the very genes that would normally build a large tree can instead build a miniature-scale model, given a suitable environment. Bonsai trees aren’t completely scaled down, of course: their cells are normal-sized – it’s just that each component is made with fewer of them.
In the 1950 and ’60s, many children were affected by their mothers taking the drug thalidomide while pregnant, when the drug blocked growth of the internal parts of their limbs. Even though growth of the skin is not directly affected by thalidomide, the very short limbs of affected children were covered by an appropriate amount of skin, not the much larger amount that would be needed to cover a normal limb. The growth of the skin cannot, therefore, just be in response to the command of a hard-wired internal blueprint: something much more adaptive must be going on.
Such observations are not troubling for biological science as such. But they are troubling for a certain picture of how biology works. The symbol for this worldview might be the DNA double helix, its complementary twisting strands evoking other interdependent pairs in life: male and female, form and function, living and non-living. DNA on its own is just a chemical polymer, after all, essential for life but not itself alive. Yet it holds out the promise that we can explain living processes purely in terms of the interactions between simple molecules.
Twentieth-century biology was dominated by a strongly reductionist agenda even before James Watson, Francis Crick, Rosalind Franklin and Maurice Wilkins revealed the structure of DNA in 1953. Early geneticists knew perfectly well that they were studying statistical correlations (if one makes such-and-such a change in gene x, then allows the organism to develop in a fixed environment e, one will observe alteration f, etc). In later generations, however, the language of causation started to creep in, and with it the idea of a fixed relationship between genes and body features. Every reader of this essay will have read or heard phrases such as ‘the gene for breast cancer’, ‘the gene for autism’ or even ‘the gene for intelligence’ (try typing any of those phrases into a search engine). The cultural idea that went with this slip of meaning – that bodies are made according to a genetic blueprint – placed genes at the centre of mainstream biological thought. The discovery and progressive unravelling of the double helix seemed only to confirm their place there.
The union of genetics with molecular biology has undoubtedly created a powerful new science. By unpicking the molecular interactions inside cells, we’ve been able to develop drugs to mimic or interfere with specific processes. The discovery of enzymes that synthesise and edit DNA laid the foundations for genetic engineering, which is usually discussed in terms of its commercial applications, but whose most common use has always been in pure research. In this way, advances in knowledge create the tools to advance knowledge, in a virtuous circle. Nevertheless, we appear to have come to a threshold. The more we know about the molecular processes, the less sense the gene-centric perspective makes.
In the early days of genetics, scientists studying different animals would identify genes and correlate them with body-scale features. However, they had no way of knowing whether the genes they had identified in one species were the same ones that were known in another. Once technologies for reading DNA sequences were invented, it became possible for researchers to ‘read’ their genes and to notice when genes in two different species were so related they were effectively the same thing.
This often produced surprises. The gene ‘Wingless’, for example, was identified by fruitfly geneticists as a blueprint for making wings (fly biologists name their genes after what happens if the gene is damaged). Cancer specialists working with mice had identified what they called Int-1 as a gene for mouse breast cancer. But these genes turned out to be effectively the same: they code for a protein now given the hybrid name Wnt. Properly analysed, Wnt is neither for making wings nor for breast cancer. It is for carrying signals between cells: the meaning of the signal depends on what cell, in what organism, is receiving it.
This is just one of very many examples. The concept of ‘the gene for feature x’ is giving way to a much more complicated story. Think something like: ‘the gene for protein a, that interacts with proteins b, c and d to allow a cell to undertake process p, that allows that cell to co‑ordinate with other cells to make body feature x’. The very length of the above phrase, and the weakness of the blueprint metaphor, emphasises a conceptual distance that is opening up between the molecular-scale, mechanical function of genes and the interesting large-scale features of bodies. The genes matter – of course they do, because something has to build all these proteins. But the helix seems less and less appropriate as an icon for the all-important control systems that run life, especially at larger scales (cells, tissues, organisms, populations, ecosystems and so on).
There is, however, an alternative. It can be represented by an even simpler icon than the double helix. It really does seem to pervade life at all scales. This alternative is a concept, rather than a physical thing. And it can it be glimpsed most clearly if we ask how things structure themselves when they must adapt to an environment that cannot be known in advance.
The most local environment for many cells consists simply of their similar neighbours. This is especially so in a very common sort of structure that we find in all sorts of living things – the sheet. (Think of the surface of the skin, the lining of the gut, the inside layer of your blood vessels.) These sheets are collections of cells stuck together, edge to edge. The cells adhere to one another via a number of very small, specialised junction complexes. The outside part of a junction complex on one cell binds to the outside part of a similar complex on the neighbouring cell, while the inside part binds to long, strong filaments of protein that form a network inside the cell: the ‘cytoskeleton’, as it is known. It is the mechanical connection of each cell’s cytoskeleton to those of its neighbours, via the junctions, that helps to give the sheet its strength.
When cells first meet, in normal development or in a culture dish, neither can ‘know’ in advance precisely where contact will first be made. The internal cytoskeletons of the cells cannot, therefore, be built to an advanced plan; they must develop adaptively to suit the precise conditions at the time. The way they achieve this is both fascinating in detail and, in its general outline, thoroughly typical of life.
The cells that are forming the sheet continuously make new branches of their cytoskeleton network. These branches head towards random points on the cell periphery. Those that do not happen to land on a junction will never experience mechanical force, whereas those filaments that do find and link to a junction will be placed under tension by the neighbouring cell. Now, it happens that cells are full of enzyme complexes that rapidly destroy cytoskeleton filaments. This destruction is, however, strongly inhibited if a filament is under mechanical load. The many filaments that end up in the wrong place are therefore quickly destroyed, while those that happen to be arranged appropriately to carry forces survive. Thus the cytoskeleton’s anatomy organises itself according to its environment and adapts continuously to changing mechanical loads.
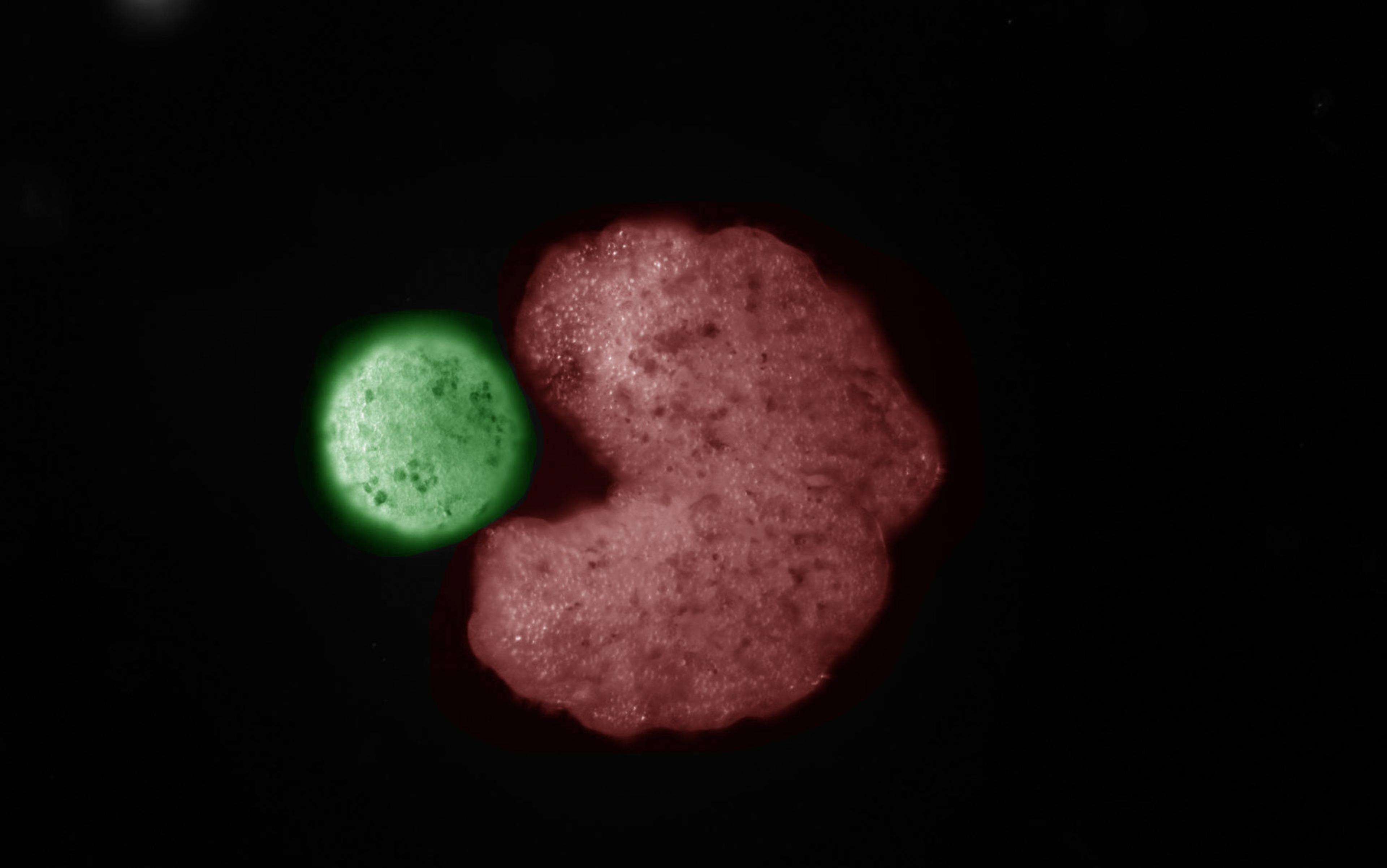
This is just a very small-scale example of adaptive self-organisation. But the same principles work at larger scales, too. Organs consist of vast numbers of cells of different types, intricately assembled to perform whatever the organ’s function might be. Some cells have a very direct connection with that function: the milk-secreting cells of the breast, for example, or the food-absorbing cells of the intestines. Others serve a support role, bringing food and oxygen, removing toxins, carrying the products of the organ wherever they need to go and so on. For the organ to work, each type of cell needs to be present in the right numbers at the right places, yet we know that organs cannot be assembled according to a precise, blueprint-like spatial plan. We know this because it is possible to remove organs from animal embryos and culture them in the presence of artificial spatial restraints, so that the overall anatomy follows the artificial constraints but the detailed internal anatomy looks normal, showing that its construction must have adapted to the new circumstances.
In some cases, we are starting to understand how this self-organisation happens, and how it relies on cell-to-cell communication. The growth of blood capillaries provides a good illustration. Tissues need blood capillaries to bring them oxygen and nutrients and to take away waste products, but a cell that is too far from the nearest blood capillary will find itself short of oxygen. At this point, a protein called HIF1A, which is normally destroyed by an oxygen-dependent process almost as soon as it is made, starts to accumulate. HIF1A puts a temporary brake on further cell proliferation and also causes the cell to secrete a protein called VEGF, which spreads out through the surrounding tissue. The cells that make blood capillary walls are sensitive to VEGF: if they detect it, they begin to proliferate and extend new capillary branches towards its source, and so the tissue cells that were short of oxygen will receive a supply from the new blood vessels. When capillary growth is adequate, there will be enough oxygen to make HIF1A unstable again, so the brake on tissue proliferation is released, VEGF production will cease and so will capillary growth. In a growing organ, this sort of thing happens again and again to make sure that growth doesn’t outstrip blood supply.
As bodies grow, they stretch the skin, which expands until the tension has been reduced: the growth of the skin keeps up with that of the body
We can tell a similar story about the fluid drainage ducts of the kidney. These ducts consist of a treelike arrangement of tubes, spread out in space to serve the organ. Even when confronted experimentally with very peculiar constraints, they will grow into a well-spaced (if strangely-shaped) tree. My laboratory has recently discovered that this spacing is achieved by, in effect, self-loathing: the growing tips of the tree’s branches secrete a molecule that they themselves find repulsive, so they grow in the direction that minimises their exposure to it and therefore maximises their mutual separation. This simple mechanism adapts naturally to ‘unexpected’ anatomical situations, whether due to experimental interventions or earlier errors in embryonic development.
Or take the skin, the largest organ of the body. It has to be exactly the right size to cover all that lies underneath, and yet the dimensions of the body cannot be predicted from the genes: variations in diet and exercise have a huge effect on body size, as does pregnancy (temporarily). Clearly, the growth of the skin has to be adaptive. A series of experiments on skin cells in culture, performed mainly by Celeste Nelson’s tissue morphodynamics group in Princeton, suggests a very elegant mechanism: skin cells respond to tension by proliferating, the daughter cells of each cell division being aligned with the tension so that the tissue grows in that direction. As bodies grow, they stretch the overlying skin, which proliferates and expands until the tension has been reduced. The growth of the skin therefore keeps up with that of the body.
If skin is damaged, it has to repair itself, adapting its activity to whatever unpredictable location and shape the hole happens to have. Adult skin heals by scarring – an ‘emergency’ response that seals the body off from a hostile world. Foetal skin, however, heals without scars. If it is cut, cells that used to have neighbours suddenly encounter free space. They respond to it by organising a contractile band along their exposed edge. This band links the cytoskeletal fibres of neighbouring cells so that the whole edge of the wound starts to contract, closing it up. As closure proceeds, cells that meet make junctions with one another and, as they no longer feel a free edge, they lose their contractile band and become normal members of a sheet again. This proceeds until the hole has disappeared completely.
Self-organising processes seem to operate at much larger scales, too – beyond the level of single organisms. Schools of fish, for example, consist of individuals that align themselves with one another so that the school moves and changes direction as a cohesive entity. This can turn a predator species into a highly efficient hunting collective – when, for example, tuna schools form outstretched arms to engulf their prey. Similar behaviour can also protect prey species: if a predator charges a school of pollock, the school will split up and reunite behind the intruder, always presenting it with a confusing swirl of hard-to-track, churning fish.
We still have much to learn about how schooling works, but it seems obvious that each fish can’t have a detailed choreography stored in its head, nor a detailed map of the location of every other fish in the school. Instead, each individual seems to base its behaviour on purely local influences: most importantly, the distances and speeds of its immediate neighbours. Computer models produce quite convincing schooling behaviour when their constituent fish – all the same, with no special leader in charge – have four basic behaviours: attraction (closing up to another fish), repulsion (increasing the distance from another fish), alignment (altering direction to swim parallel to a neighbour) and searching (looking for a school of fish). Searching happens only if a fish finds itself isolated. Repulsion is strong at short distances and attraction kicks in at larger ones, which tends to keep fish an optimal distance apart, and they reorientate to swim in the average direction of their various neighbours.
large-scale ecosystems of co‑operating organisms such as trees and fungi show self‑organising behaviour
These simple rules are not enough to mimic the particulars of hunting or predator-evading behaviour, but they are enough to keep a school organised for its routine swimming. The flocking of birds appears to work in a similar way: almost 30 years ago, the programmer Craig Reynolds made a computer model of very simple entities – ‘boids’ – that also showed attraction, repulsion, and alignment. Again, the behaviour of the model is so good it implies that real birds might well organise their flocks by each obeying simple, local rules.
Schooling and flocking behaviours are, of course, restricted to large groups of individuals from a single species. Might self-organising properties operate even at the level of multi-species ecosystems? There’s reason to think that they do. Vegetation on arid soils will, for example, arrange itself into groups with plain soil between them: the plants in the groups all benefit from their mutual ability to help rare rainfall penetrate the ground. And in simple experimental microbial communities (not, in fact, different species, but yeasts genetically engineered to have different metabolisms that can co-operate to use environmental nutrients), the different types of individual self-organise into mixed clusters that bring co‑operators together. Many ecologists believe that large-scale ecosystems of co‑operating organisms, for example trees and fungi, show similar self‑organising behaviour.
All of these examples, and many more like them, turn out to have something in common when analysed at the mechanistic level: in each case, what has been achieved so far by the system is used to control its current behaviour. This type of control is called feedback, and is represented by a loop feeding information from the output of a process back to its input.
In the case of the cytoskeleton, the stability of a filament depended on whether it was carrying a mechanical force, which in turn depended on whether it was in the right place to connect to cell junctions. The achievement of a filament (to be in a useful place or not) is therefore fed back to decide what it will do next (survive, or be disassembled). In the case of the blood capillaries, the extent to which present growth has been adequate to bring enough oxygen into the tissues is fed back, via VEGF, to control whether the capillaries continue to grow or remain as they are. And the same principle seemed to explain the schooling of fish: any error in an individual fish’s relative positions and direction compared to its neighbours is used to modify its swimming, to make the error smaller. Seen from the abstract perspective of feedback loops, adaptive self-organisation looks more or less the same across all scales of life, from the architecture of subcellular assemblies to the arrangements of co-operating species in ecosystems.
In this sense, the loop is a near-universal symbol of living processes.
We have, then, two very different models for understanding life. In the gene-centred model, a ‘gene for this’ and a ‘gene for that’ act deterministically to construct a cell, or a body, or an aspect of behaviour. In the loop-centred model, feedback-rich processes allow cells, bodies and ecosystems to construct themselves adaptively in response to the prevailing conditions. Can these models, which seem on the face of it very different, ever be reconciled? Yes, very easily. The feedback loops that guide self-organisation, at any scale, rely ultimately on the action of protein-based mechanisms, and proteins are encoded by genes.
genes do not make body features, they make the machines that organise body features adaptively
The role of proteins in subcellular self-organising systems such as the cytoskeleton is obvious: the cytoskeleton is made of proteins. In my account of capillary development, the proteins HIF1A and VEGF highlighted the importance of specific molecules to organisation at a larger scale. Even events at much larger scales, such as the schooling of fish, ultimately rely on protein machines. Genes are therefore essential to self-organisation at all the scales of life – just not in a deterministic way. Rather, the genes are needed to make the machines that mediate feedback-driven self-organisation: the self-organisation is a high-level property that emerges from the underlying network, not a feature of any of the individual components.
This has interesting consequences. Where any part of the mechanism is sensitive to the environment, the whole self-organising loop can be too. The number of red cells in blood, for example, is set by a feedback loop sensitive to measurements of oxygen deep in the kidneys. When someone goes to live in the thin air of a high mountain, they tend to have more blood cells. Why? Because the ‘normal’ complement of cells is not sufficient in that environment, so the kidneys sense abnormally low oxygen and signal for more blood cells to be made. The effects of environmental sensitivity at a single point percolate throughout the entire organism. If we recognise that genes do not make body features, they make the machines that organise body features adaptively, that shift in perspective does much to lay to rest the long debates about nature versus nurture.
The DNA helix is important, of course. But the most important thing it does is make proteins that can operate in regulatory loops. These loops can also operate at the molecular level: genes make proteins, but these proteins determine which genes are ‘off’ and which are ‘on’ (as HIF1A does), making a control loop at even the molecular level. Unlike the helix, loops also operate at scales far above the molecular, covering a range of sizes from bacterial colonies to the vast ecosystems of the rainforest – perhaps to the ecosystem of the entire Earth. Beyond Earth, life without DNA is just about thinkable (one can imagine alternative strategies for storing information). Life without feedback loops, though? I have never met any biologist who can imagine that.
The helix is too well-established an icon to be deposed any time soon. And yet, a simple loop would be a much more universal symbol of how life works at all of its scales and levels. Perhaps the Ouroboros, beloved of gnostics and alchemists, has been an ideal symbol waiting in the wings for centuries: there can surely be no more evocative symbol of feedback than a snake growing by devouring its own tail.